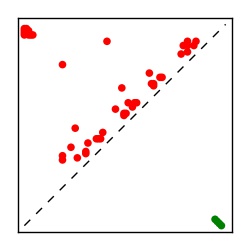
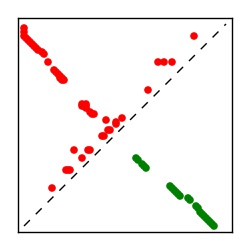
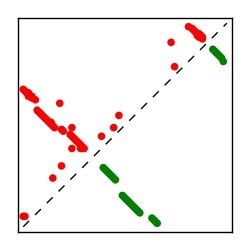
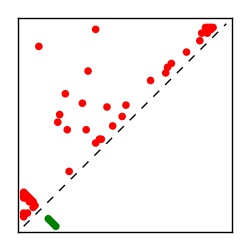
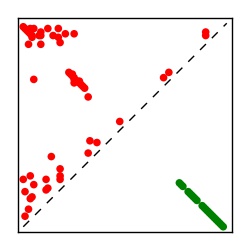
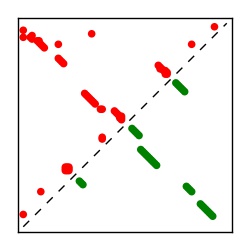
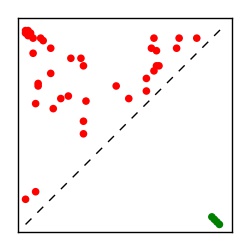
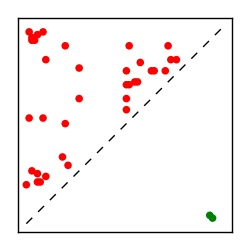
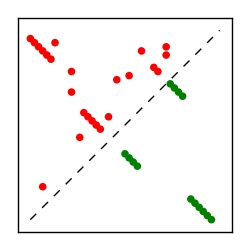
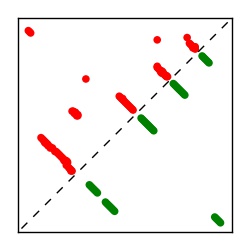
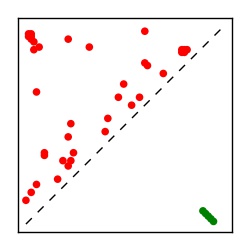
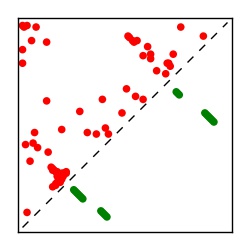
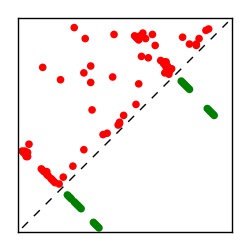
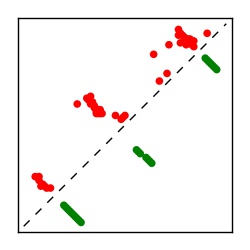
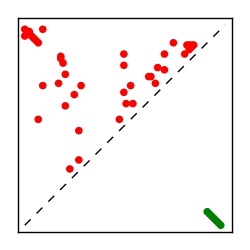
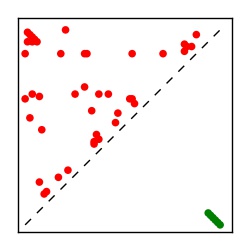

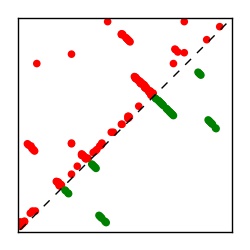
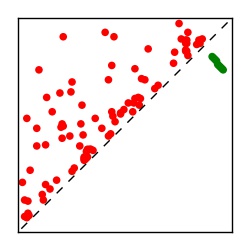
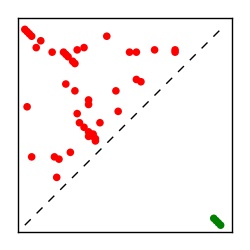
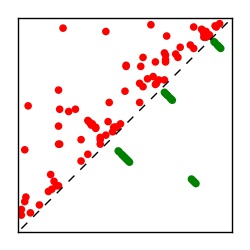
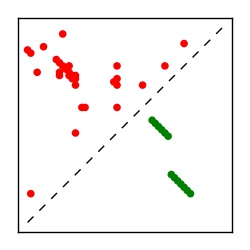
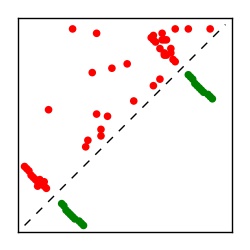
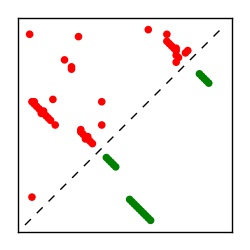
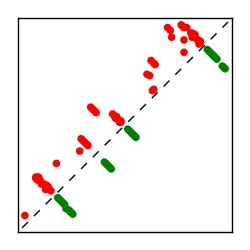
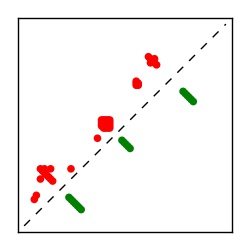
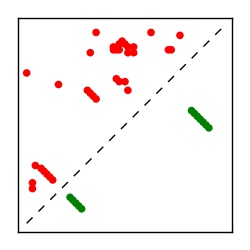
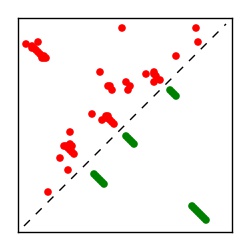
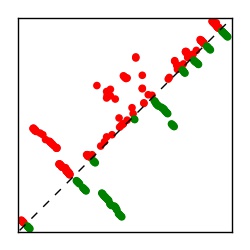
Supplementary Table 1. Summary of 182 RFAM families with EC predictions
Supplementary Table 2. Mapping to RFAM reduced numbering
Supplementary Table 3. Mapping from RFAM reduced numbering to PDB numbering
Supplementary Table 4. Annotated lists of contacts from 22 RFAM families.
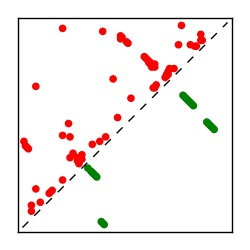
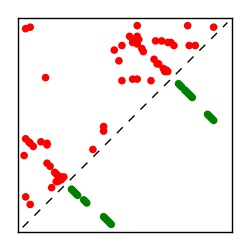
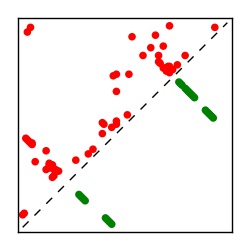
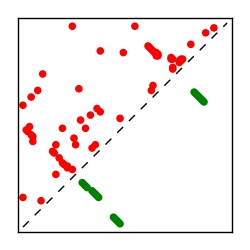
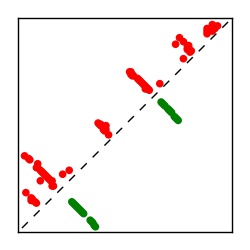
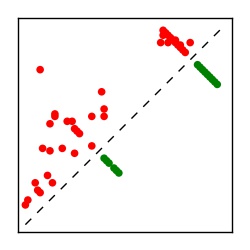
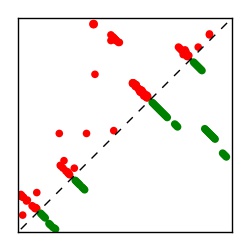
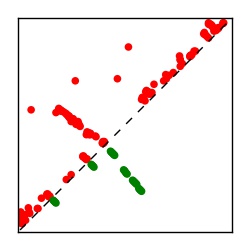
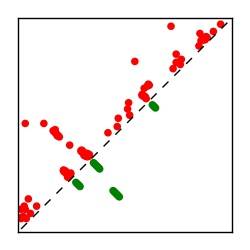
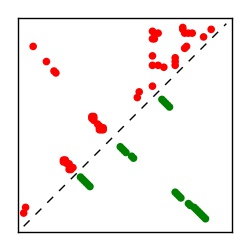
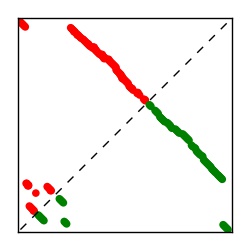
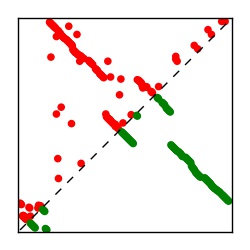
Here we provide data on evolutionary couplings (ECs) for 182 RNA families from RFAM, computed using a global maximum entropy probability model that is available for download here. Each EC score represents the strength of co-variation between two RNA nucleotides and can be used to infer 3D contacts in the folded RNA. Before computing ECs for an RFAM alignment, we remove all columns with > 50% gaps, producing an RFAM reduced mapping that maps the the original RFAM numbering to our new numbering with highly gapped columns removed. 22 of the 182 RFAM alignments have a PDB structure for one member of the family. For each of these, we created a PDB mapping that maps our column-reduced alignment to the PDB structure. Below, we present validation data on the 22 families with a structure, as well as predicted contacts for 160 RFAM families without a known structure. Some if this data is also contained in the supplementary tables (above) from the paper. Table column names are defined below.
| Column name | Description |
|---|---|
| L | Length of the reduced alignment (i.e. number of columns) |
| M | Total number of sequences in the alignment |
| Meff | Effective number of sequences in the alignment (similar sequences are down-weighted) |
| RFAM reduced mapping | Maps RFAM numbering to our numbering where highly gapped columns have been removed |
| PDB mapping | Maps our numbering (RFAM reduced) to PDB numbering |
| Top ECs (annotated) / EC file | List of nucleotide pairs ranked by EC score, annotated when a PDB structure is available |
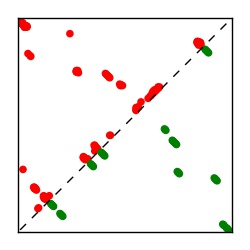
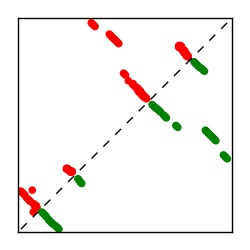
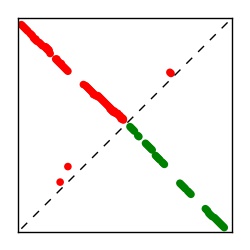
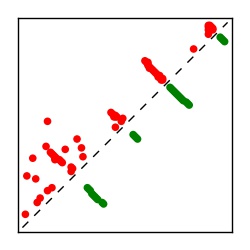
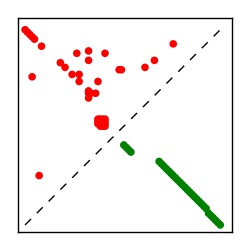
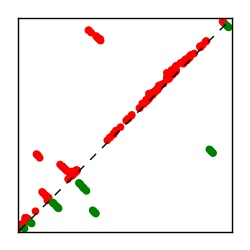
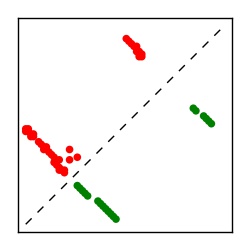
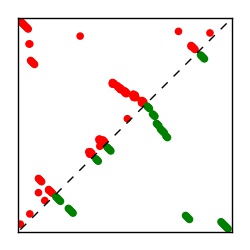
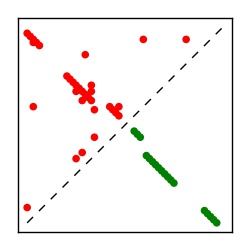
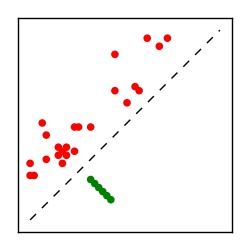
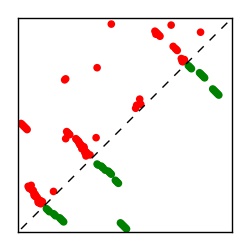
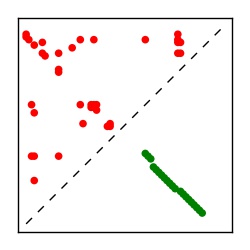
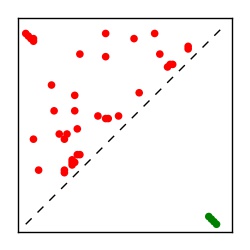
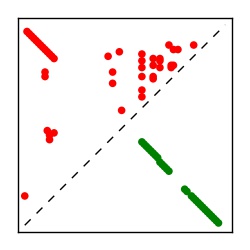
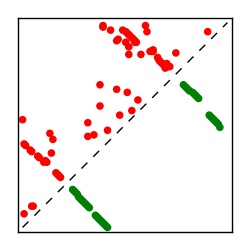
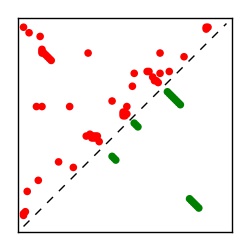
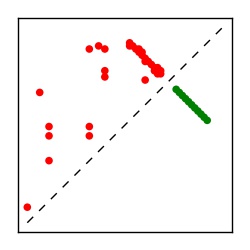
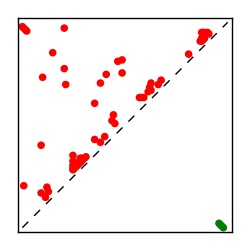
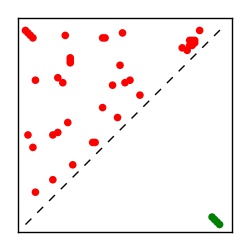
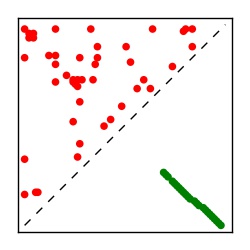
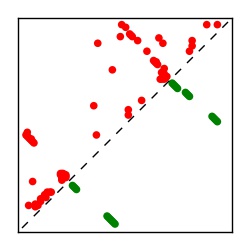
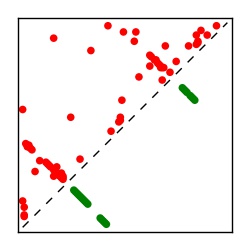
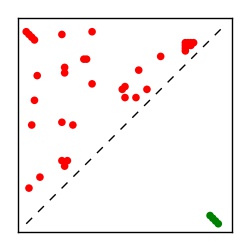
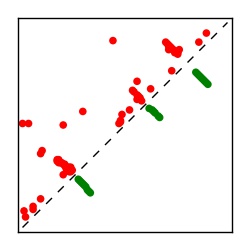
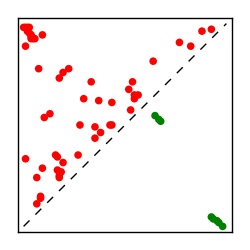
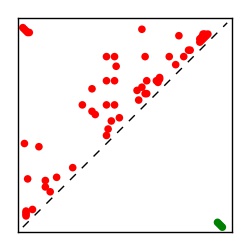
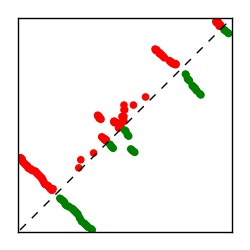
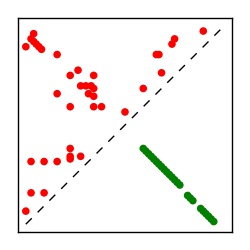
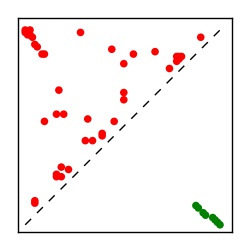
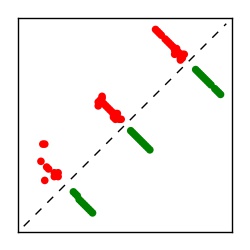
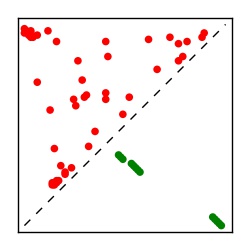
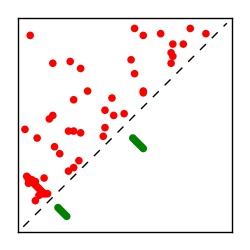
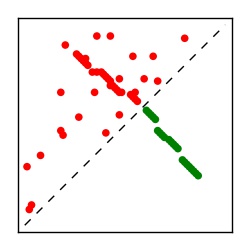
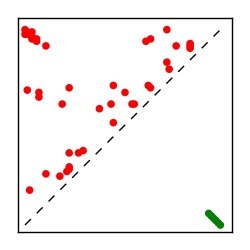
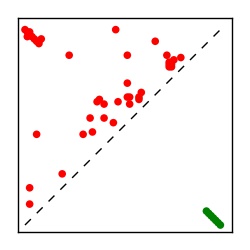
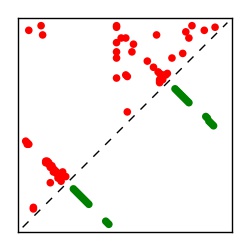
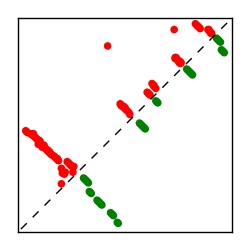
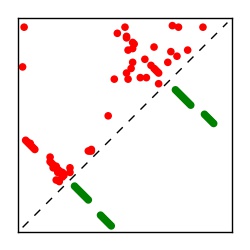
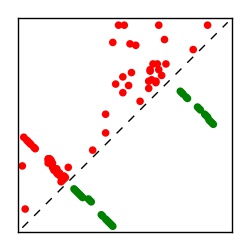
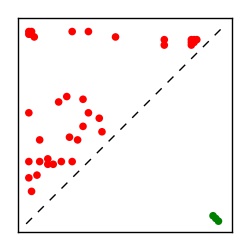
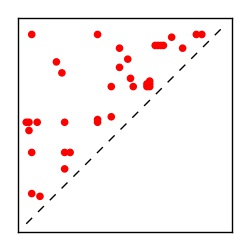
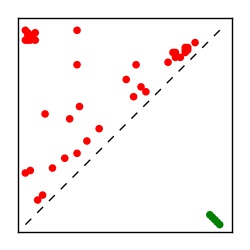
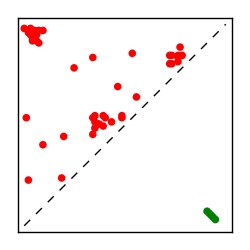
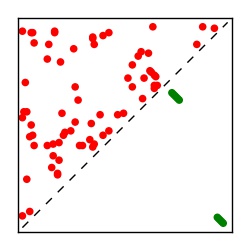
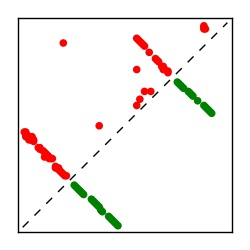
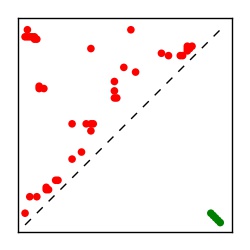
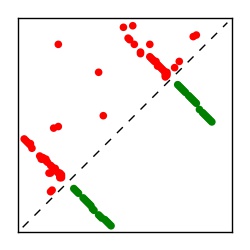
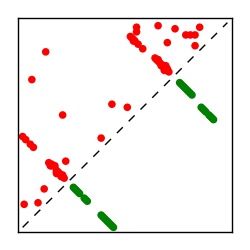
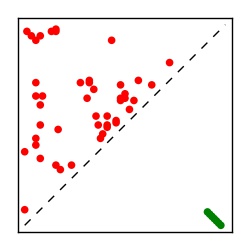
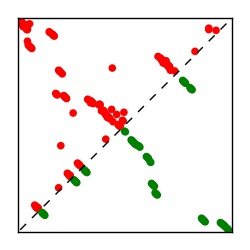
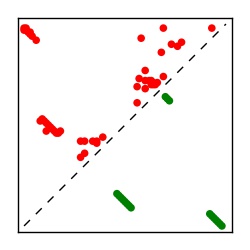
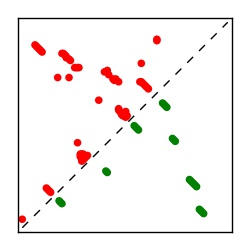
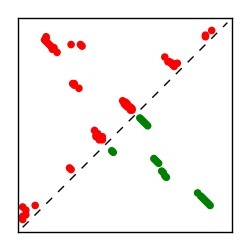
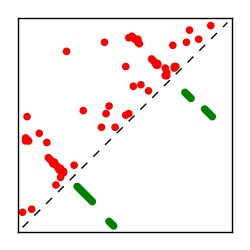
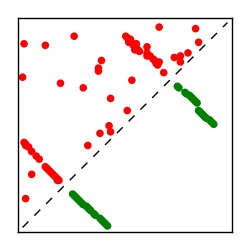
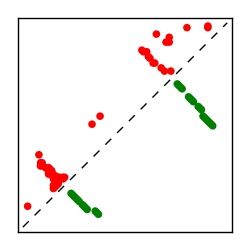
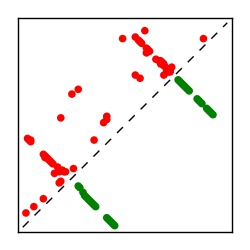
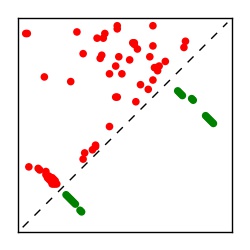
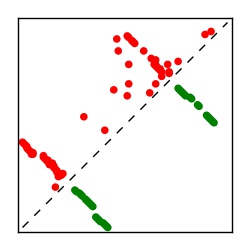
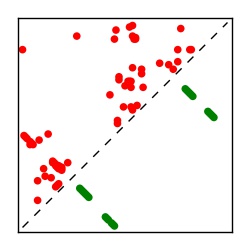
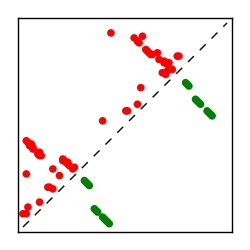
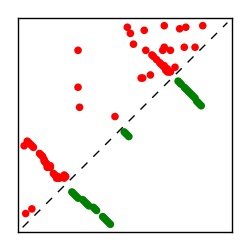
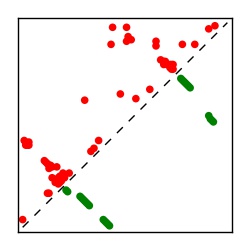
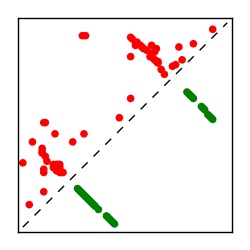
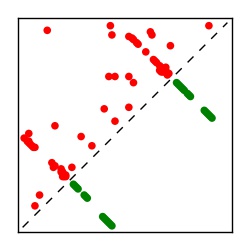
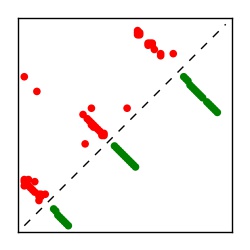
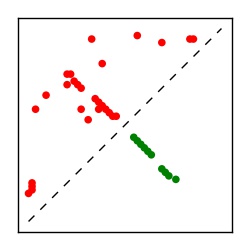
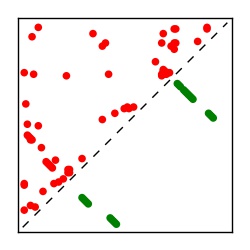
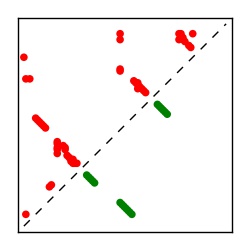
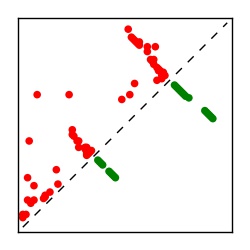
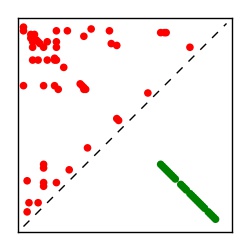
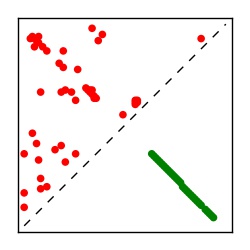
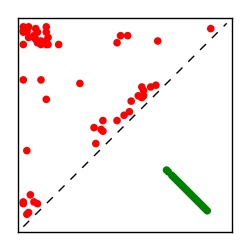
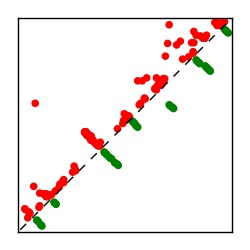
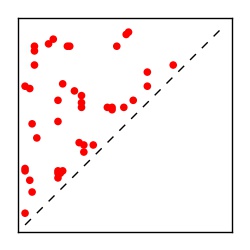
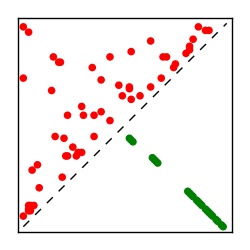
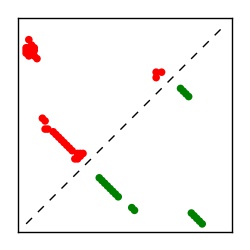
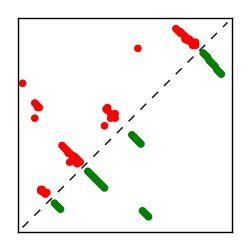
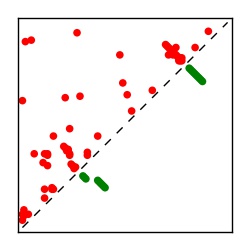
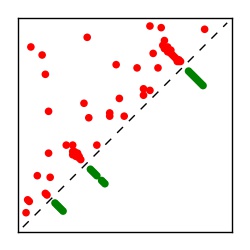
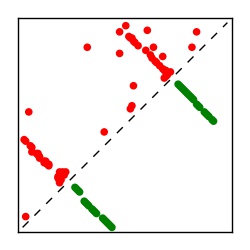
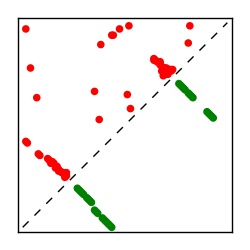
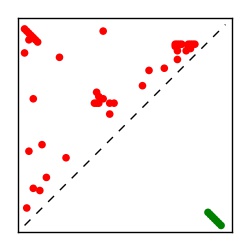
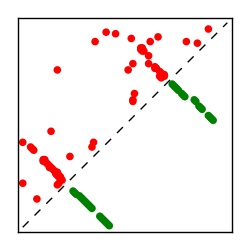
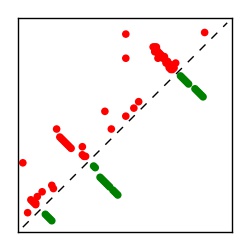
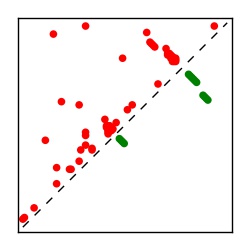
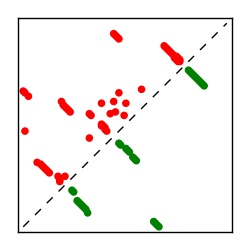
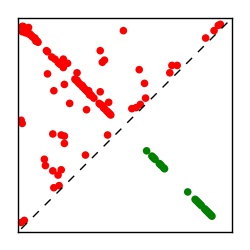
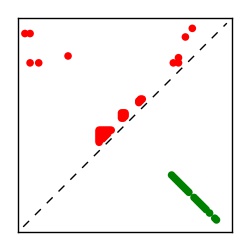
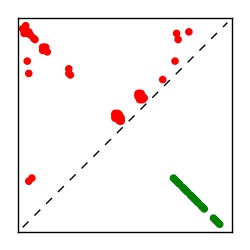
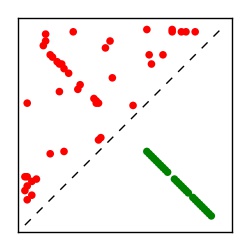
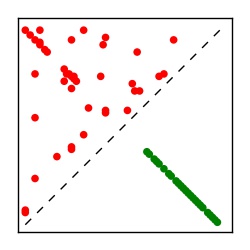
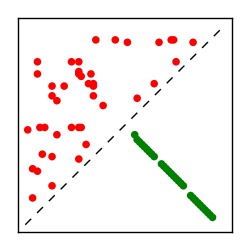
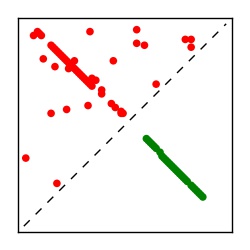
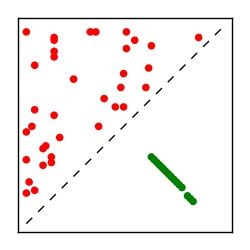
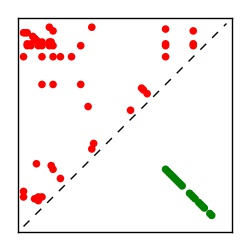
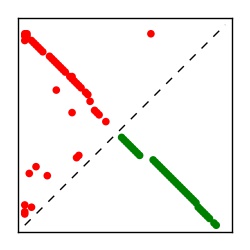
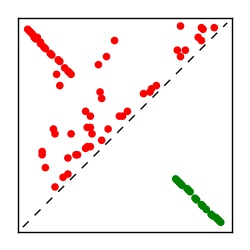
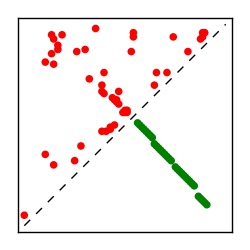
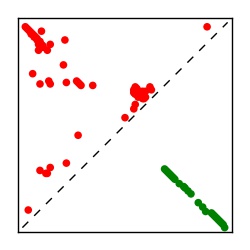
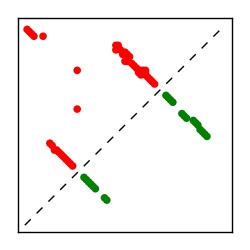
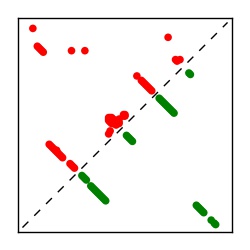
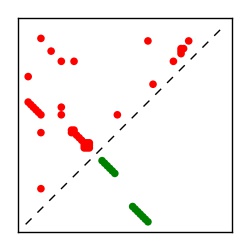
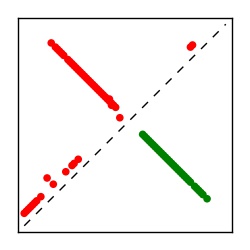
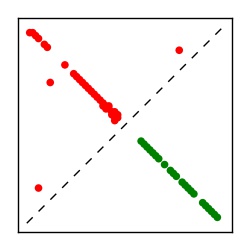
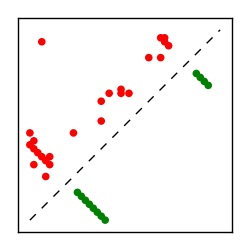
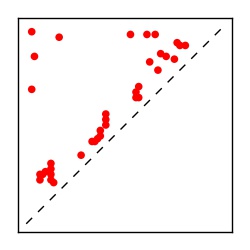
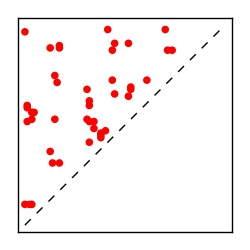
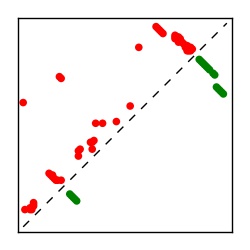
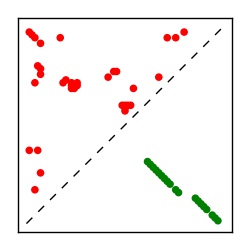
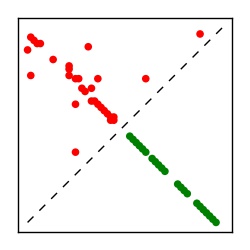
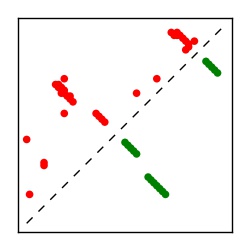
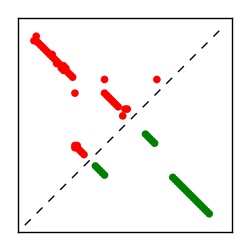
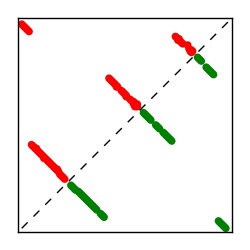
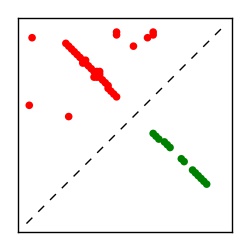
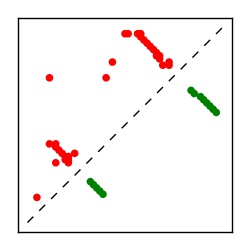
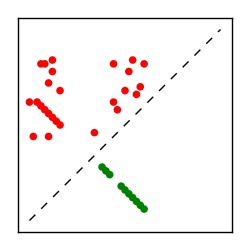
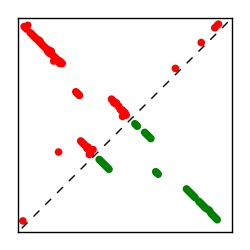
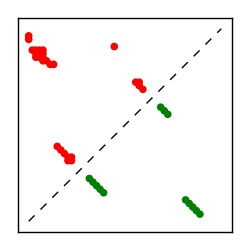
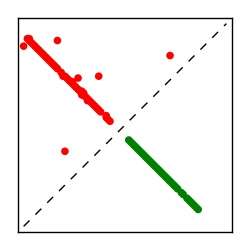
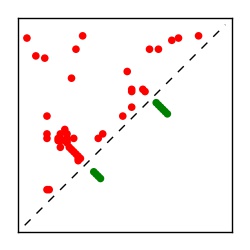
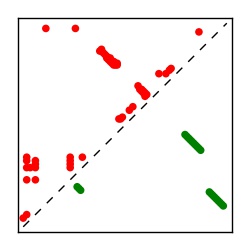
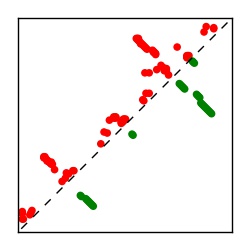
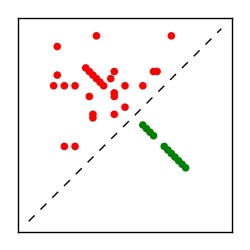
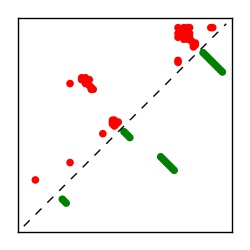
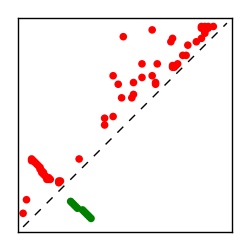
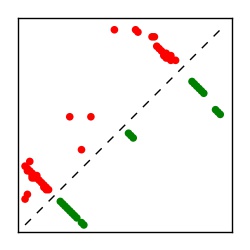
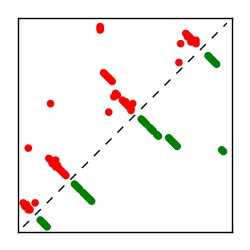
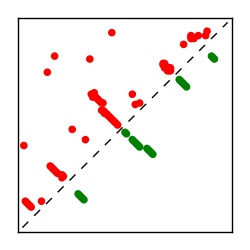
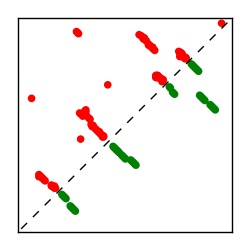
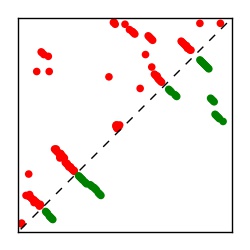
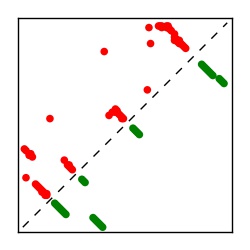
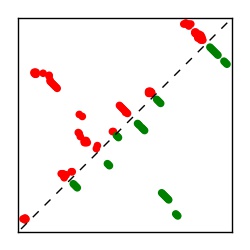
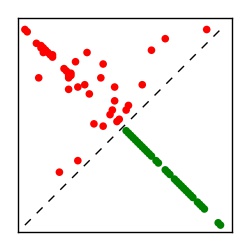
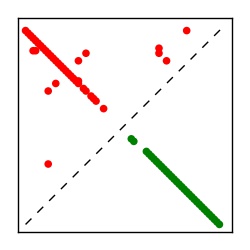
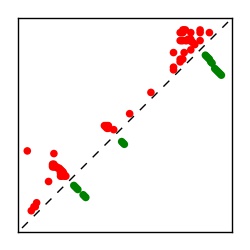
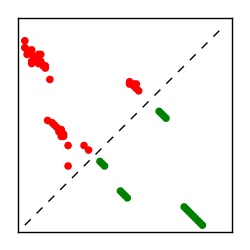
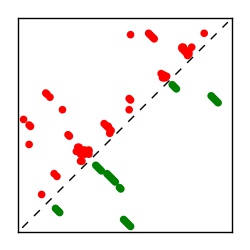
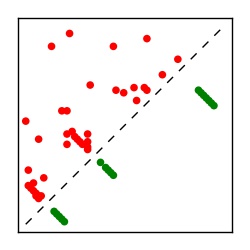
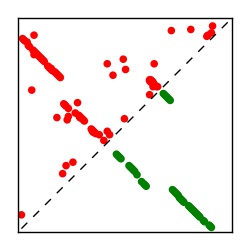
| Contact map | Figure showing top ranking ECs (tdumbnails link to high resolution pdf) |
| TP curve | Figure showing mean true positive rate versus number of contacts |
| RFAM ID | Name | L | M | Meff | RFAM reduced mapping | EC file | Contact map |
|---|---|---|---|---|---|---|---|
| RF00003 | U1 spliceosomal RNA | 165 | 16344 | 2356.3 | Reduced aligment | ECs |  |
| RF00004 | U2 spliceosomal RNA | 192 | 11870 | 1930.5 | Reduced aligment | ECs |  |
| RF00006 | Vault RNA | 101 | 600 | 173.1 | Reduced aligment | ECs |  |
| RF00007 | U12 minor spliceosomal RNA | 151 | 539 | 106.2 | Reduced aligment | ECs |  |
| RF00009 | Nuclear RNase P | 295 | 895 | 370.5 | Reduced aligment | ECs |  |
| RF00012 | Small nucleolar RNA U3 | 219 | 3110 | 477.4 | Reduced aligment | ECs |  |
| RF00013 | 6S / SsrS RNA | 187 | 3521 | 648.2 | Reduced aligment | ECs |  |
| RF00015 | U4 spliceosomal RNA | 139 | 9417 | 2328.9 | Reduced aligment | ECs |  |
| RF00016 | Small nucleolar RNA SNORD14 | 95 | 1672 | 203.6 | Reduced aligment | ECs |  |
| RF00019 | Y RNA | 101 | 22242 | 1192.6 | Reduced aligment | ECs |  |
| RF00020 | U5 spliceosomal RNA | 117 | 5700 | 530.7 | Reduced aligment | ECs |  |
| RF00026 | U6 spliceosomal RNA | 107 | 72126 | 6991.8 | Reduced aligment | ECs |  |
| RF00027 | let-7 microRNA precursor | 85 | 1263 | 57.5 | Reduced aligment | ECs |  |
| RF00028 | Group I catalytic intron | 447 | 59999 | 2087.7 | Reduced aligment | ECs |  |
| RF00029 | Group II catalytic intron | 77 | 51464 | 1297.7 | Reduced aligment | ECs |  |
| RF00030 | RNase MRP | 266 | 424 | 170.4 | Reduced aligment | ECs |  |
| RF00031 | Selenocysteine insertion sequence 1 | 64 | 1278 | 60.6 | Reduced aligment | ECs |  |
| RF00032 | Histone 3' UTR stem-loop | 47 | 11099 | 550.5 | Reduced aligment | ECs |  |
| RF00045 | Small nucleolar RNA SNORA73 family | 205 | 1032 | 237.3 | Reduced aligment | ECs |  |
| RF00047 | mir-2 microRNA precursor | 73 | 381 | 63.8 | Reduced aligment | ECs |  |
| RF00049 | Small nucleolar RNA SNORD36 | 77 | 597 | 77.3 | Reduced aligment | ECs |  |
| RF00053 | mir-7 microRNA precursor | 89 | 348 | 48.5 | Reduced aligment | ECs |  |
| RF00056 | Small nucleolar RNA SNORA71 | 135 | 613 | 100.8 | Reduced aligment | ECs |  |
| RF00062 | HgcC family RNA | 110 | 154 | 115.3 | Reduced aligment | ECs |  |
| RF00066 | U7 small nuclear RNA | 63 | 5881 | 134.8 | Reduced aligment | ECs |  |
| RF00067 | Small nucleolar RNA SNORD15 | 142 | 477 | 82.4 | Reduced aligment | ECs |  |
| RF00069 | Small nucleolar RNA SNORD24 | 80 | 536 | 81.1 | Reduced aligment | ECs |  |
| RF00073 | mir-156 microRNA precursor | 91 | 789 | 55.5 | Reduced aligment | ECs |  |
| RF00075 | mir-166 microRNA precursor | 116 | 1106 | 201.7 | Reduced aligment | ECs |  |
| RF00080 | yybP-ykoY leader | 116 | 1882 | 206.5 | Reduced aligment | ECs |  |
| RF00086 | Small nucleolar RNA SNORD27 | 79 | 291 | 47.3 | Reduced aligment | ECs |  |
| RF00089 | Small nucleolar RNA SNORD31 | 71 | 280 | 50.6 | Reduced aligment | ECs |  |
| RF00091 | Small nucleolar RNA SNORA62/SNORA6 family | 155 | 714 | 181.2 | Reduced aligment | ECs |  |
| RF00092 | Small nucleolar RNA SNORA63 | 132 | 712 | 130.7 | Reduced aligment | ECs |  |
| RF00093 | Small nucleolar RNA SNORD18 | 72 | 576 | 91.3 | Reduced aligment | ECs |  |
| RF00096 | U8 small nucleolar RNA | 136 | 794 | 148.1 | Reduced aligment | ECs |  |
| RF00097 | Plant small nucleolar RNA R71 | 108 | 11540 | 466.6 | Reduced aligment | ECs |  |
| RF00099 | Small nucleolar RNA SNORD22 | 127 | 288 | 63.6 | Reduced aligment | ECs |  |
| RF00100 | 7SK RNA | 311 | 21885 | 8715.2 | Reduced aligment | ECs |  |
| RF00104 | mir-10 microRNA precursor family | 76 | 637 | 40.6 | Reduced aligment | ECs |  |
| RF00105 | Small nucleolar RNA SNORD115 | 83 | 2565 | 107.2 | Reduced aligment | ECs |  |
| RF00106 | RNAI | 106 | 1687 | 67.3 | Reduced aligment | ECs |  |
| RF00108 | Small nucleolar RNA SNORD116 | 94 | 1854 | 111.6 | Reduced aligment | ECs |  |
| RF00114 | Ribosomal S15 leader | 117 | 1082 | 110.3 | Reduced aligment | ECs |  |
| RF00127 | t44 RNA | 89 | 1075 | 109.9 | Reduced aligment | ECs |  |
| RF00133 | Small nucleolar RNA Z195/SNORD33/SNORD32 family | 86 | 618 | 138.8 | Reduced aligment | ECs |  |
| RF00134 | Small nucleolar RNA Z196/R39/R59 family | 86 | 418 | 99.8 | Reduced aligment | ECs |  |
| RF00139 | Small nucleolar RNA SNORA72 | 133 | 548 | 102.8 | Reduced aligment | ECs |  |
| RF00140 | Alpha operon ribosome binding site | 111 | 1226 | 93.7 | Reduced aligment | ECs |  |
| RF00147 | Small nucleolar RNA SNORD34 | 80 | 202 | 45.0 | Reduced aligment | ECs |  |
| RF00155 | Small nucleolar RNA SNORA66 | 134 | 246 | 79.7 | Reduced aligment | ECs |  |
| RF00156 | Small nucleolar RNA SNORA70 | 136 | 2128 | 499.5 | Reduced aligment | ECs |  |
| RF00163 | Hammerhead ribozyme (type I) | 46 | 49036 | 54.5 | Reduced aligment | ECs |  |
| RF00168 | Lysine riboswitch | 180 | 2422 | 439.7 | Reduced aligment | ECs |  |
| RF00181 | Small nucleolar RNA SNORD113/SNORD114 family | 75 | 2257 | 251.9 | Reduced aligment | ECs |  |
| RF00190 | Small nucleolar RNA SNORA16B/SNORA16A family | 136 | 331 | 82.7 | Reduced aligment | ECs |  |
| RF00191 | Small nucleolar RNA SNORA57 | 150 | 336 | 110.4 | Reduced aligment | ECs |  |
| RF00198 | SL1 RNA | 106 | 1802 | 105.2 | Reduced aligment | ECs |  |
| RF00213 | Small nucleolar RNA R38 | 89 | 617 | 103.9 | Reduced aligment | ECs |  |
| RF00218 | Small nucleolar RNA SNORD46 | 84 | 365 | 74.4 | Reduced aligment | ECs |  |
| RF00230 | T-box leader | 216 | 16195 | 3389.8 | Reduced aligment | ECs |  |
| RF00263 | Small nucleolar RNA SNORA68 | 134 | 388 | 114.1 | Reduced aligment | ECs |  |
| RF00272 | Small nucleolar RNA SNORA67 | 143 | 352 | 79.7 | Reduced aligment | ECs |  |
| RF00274 | Small nucleolar RNA SNORD57 | 73 | 137 | 42.6 | Reduced aligment | ECs |  |
| RF00277 | Small nucleolar RNA SNORD49 | 72 | 196 | 38.9 | Reduced aligment | ECs |  |
| RF00284 | Small nucleolar RNA SNORD74 | 81 | 283 | 53.7 | Reduced aligment | ECs |  |
| RF00309 | Small nucleolar RNA snR60/Z15/Z230/Z193/J17 | 97 | 517 | 156.7 | Reduced aligment | ECs |  |
| RF00321 | Small nucleolar RNA Z247 | 140 | 506 | 145.4 | Reduced aligment | ECs |  |
| RF00322 | Small nucleolar RNA SNORA31 | 131 | 1102 | 223.1 | Reduced aligment | ECs |  |
| RF00324 | Small nucleolar RNA MBII-202 | 85 | 187 | 43.5 | Reduced aligment | ECs |  |
| RF00334 | Small nucleolar RNA SNORA3/SNORA45 family | 132 | 533 | 95.2 | Reduced aligment | ECs |  |
| RF00340 | Small nucleolar RNA SNORA36 family | 133 | 454 | 98.1 | Reduced aligment | ECs |  |
| RF00343 | Small nucleolar RNA Z122 | 90 | 219 | 48.3 | Reduced aligment | ECs |  |
| RF00373 | Archaeal RNase P | 289 | 463 | 168.4 | Reduced aligment | ECs |  |
| RF00375 | HIV primer binding site (PBS) | 100 | 4343 | 128.0 | Reduced aligment | ECs |  |
| RF00379 | ydaO/yuaA leader | 145 | 1656 | 531.1 | Reduced aligment | ECs |  |
| RF00391 | RtT RNA | 131 | 2746 | 108.2 | Reduced aligment | ECs |  |
| RF00392 | Small nucleolar RNA SNORA5 | 135 | 431 | 109.3 | Reduced aligment | ECs |  |
| RF00396 | Small nucleolar RNA SNORA13 | 135 | 227 | 80.1 | Reduced aligment | ECs |  |
| RF00400 | Small nucleolar RNA SNORA28 | 127 | 231 | 64.9 | Reduced aligment | ECs |  |
| RF00402 | Small nucleolar RNA SNORA25 | 129 | 833 | 196.7 | Reduced aligment | ECs |  |
| RF00405 | Small nucleolar RNA SNORA44 | 132 | 414 | 89.1 | Reduced aligment | ECs |  |
| RF00406 | Small nucleolar RNA SNORA42/SNORA80 family | 137 | 1074 | 276.0 | Reduced aligment | ECs |  |
| RF00408 | Small nucleolar RNA SNORA1 | 136 | 400 | 105.3 | Reduced aligment | ECs |  |
| RF00409 | Small nucleolar RNA SNORA7 | 139 | 26339 | 1327.9 | Reduced aligment | ECs |  |
| RF00410 | Small nucleolar RNA SNORA2/SNORA34 family | 138 | 1401 | 114.2 | Reduced aligment | ECs |  |
| RF00411 | Small nucleolar RNA SNORA9 | 134 | 431 | 91.4 | Reduced aligment | ECs |  |
| RF00413 | Small nucleolar RNA SNORA19 | 131 | 521 | 142.0 | Reduced aligment | ECs |  |
| RF00420 | Small nucleolar RNA SNORA61 | 124 | 1271 | 785.4 | Reduced aligment | ECs |  |
| RF00421 | Small nucleolar RNA SNORA32 | 123 | 540 | 77.0 | Reduced aligment | ECs |  |
| RF00425 | Small nucleolar RNA SNORA18 | 133 | 623 | 132.8 | Reduced aligment | ECs |  |
| RF00428 | Small nucleolar RNA SNORA38 | 132 | 398 | 67.9 | Reduced aligment | ECs |  |
| RF00430 | Small nucleolar RNA SNORA54 | 127 | 191 | 69.2 | Reduced aligment | ECs |  |
| RF00432 | Small nucleolar RNA SNORA51 | 133 | 844 | 182.5 | Reduced aligment | ECs |  |
| RF00435 | Repression of heat shock gene expression (ROSE) element | 96 | 111 | 49.9 | Reduced aligment | ECs |  |
| RF00436 | UnaL2 LINE 3' element | 55 | 100501 | 1719.0 | Reduced aligment | ECs |  |
| RF00438 | Small nucleolar RNA SNORA33 | 131 | 398 | 93.3 | Reduced aligment | ECs |  |
| RF00442 | ykkC-yxkD leader | 103 | 741 | 220.5 | Reduced aligment | ECs |  |
| RF00443 | Small nucleolar RNA SNORA27 | 128 | 377 | 79.0 | Reduced aligment | ECs |  |
| RF00445 | mir-399 microRNA precursor family | 111 | 490 | 160.8 | Reduced aligment | ECs |  |
| RF00451 | mir-395 microRNA precursor family | 98 | 1549 | 141.3 | Reduced aligment | ECs |  |
| RF00452 | mir-172 microRNA precursor family | 115 | 609 | 114.0 | Reduced aligment | ECs |  |
| RF00459 | Mason-Pfizer monkey virus packaging signal | 280 | 740 | 183.8 | Reduced aligment | ECs |  |
| RF00476 | Small nucleolar RNA snR61/Z1/Z11 | 85 | 341 | 87.3 | Reduced aligment | ECs |  |
| RF00485 | Potassium channel RNA editing signal | 115 | 3411 | 516.7 | Reduced aligment | ECs |  |
| RF00513 | Tryptophan operon leader | 98 | 704 | 53.6 | Reduced aligment | ECs |  |
| RF00515 | PyrR binding site | 114 | 1790 | 288.1 | Reduced aligment | ECs |  |
| RF00519 | suhB | 78 | 535 | 228.6 | Reduced aligment | ECs |  |
| RF00521 | SAM riboswitch (alpha-proteobacteria) | 80 | 356 | 99.8 | Reduced aligment | ECs |  |
| RF00525 | Flavivirus DB element | 73 | 6950 | 40.7 | Reduced aligment | ECs |  |
| RF00548 | U11 spliceosomal RNA | 135 | 677 | 158.6 | Reduced aligment | ECs |  |
| RF00557 | Ribosomal protein L10 leader | 139 | 1398 | 288.6 | Reduced aligment | ECs |  |
| RF00558 | Ribosomal protein L20 leader | 127 | 1132 | 153.1 | Reduced aligment | ECs |  |
| RF00559 | Ribosomal protein L21 leader | 78 | 891 | 61.8 | Reduced aligment | ECs |  |
| RF00560 | Small nucleolar RNA SNORA17 | 133 | 5155 | 618.2 | Reduced aligment | ECs |  |
| RF00561 | Small nucleolar RNA SNORA40 | 129 | 1001 | 159.0 | Reduced aligment | ECs |  |
| RF00568 | Small nucleolar RNA SNORA26 | 123 | 586 | 113.4 | Reduced aligment | ECs |  |
| RF00598 | Small nucleolar RNA SNORA76 | 133 | 343 | 89.4 | Reduced aligment | ECs |  |
| RF00601 | Small Cajal body specific RNA 20 | 131 | 302 | 70.1 | Reduced aligment | ECs |  |
| RF00604 | Small nucleolar RNA SNORD88 | 92 | 296 | 51.1 | Reduced aligment | ECs |  |
| RF00614 | Small nucleolar RNA SNORA11 | 130 | 719 | 111.0 | Reduced aligment | ECs |  |
| RF00618 | U4atac minor spliceosomal RNA | 127 | 715 | 124.9 | Reduced aligment | ECs |  |
| RF00619 | U6atac minor spliceosomal RNA | 127 | 2585 | 574.0 | Reduced aligment | ECs |  |
| RF00634 | S-adenosyl methionine (SAM) riboswitch, | 117 | 468 | 79.1 | Reduced aligment | ECs |  |
| RF00638 | microRNA MIR159 | 197 | 853 | 159.4 | Reduced aligment | ECs |  |
| RF00640 | microRNA MIR167_1 | 122 | 750 | 99.7 | Reduced aligment | ECs |  |
| RF00641 | microRNA mir-154 | 81 | 1954 | 66.0 | Reduced aligment | ECs |  |
| RF00643 | microRNA MIR171_1 | 103 | 568 | 137.7 | Reduced aligment | ECs |  |
| RF00645 | microRNA MIR169_2 | 118 | 880 | 348.2 | Reduced aligment | ECs |  |
| RF00647 | microRNA MIR164 | 118 | 579 | 63.9 | Reduced aligment | ECs |  |
| RF00648 | microRNA MIR396 | 133 | 390 | 87.2 | Reduced aligment | ECs |  |
| RF00655 | microRNA mir-28 | 87 | 633 | 258.4 | Reduced aligment | ECs |  |
| RF00665 | microRNA mir-290 | 82 | 554 | 51.2 | Reduced aligment | ECs |  |
| RF00670 | microRNA mir-105 | 82 | 308 | 45.5 | Reduced aligment | ECs |  |
| RF00685 | microRNA mir-36 | 104 | 263 | 76.1 | Reduced aligment | ECs |  |
| RF00693 | microRNA mir-147 | 71 | 314 | 40.6 | Reduced aligment | ECs |  |
| RF00695 | microRNA MIR398 | 110 | 244 | 58.2 | Reduced aligment | ECs |  |
| RF00697 | microRNA mir-186 | 94 | 538 | 81.2 | Reduced aligment | ECs |  |
| RF00711 | microRNA mir-449 | 91 | 294 | 73.0 | Reduced aligment | ECs |  |
| RF00736 | microRNA mir-320 | 83 | 541 | 87.7 | Reduced aligment | ECs |  |
| RF00761 | microRNA mir-340 | 98 | 967 | 266.6 | Reduced aligment | ECs |  |
| RF00779 | microRNA MIR474 | 89 | 222 | 147.7 | Reduced aligment | ECs |  |
| RF00782 | microRNA MIR480 | 127 | 586 | 372.9 | Reduced aligment | ECs |  |
| RF00825 | microRNA mir-344 | 96 | 471 | 70.7 | Reduced aligment | ECs |  |
| RF00865 | microRNA MIR169_5 | 140 | 476 | 188.2 | Reduced aligment | ECs |  |
| RF00872 | microRNA mir-652 | 97 | 656 | 175.4 | Reduced aligment | ECs |  |
| RF00875 | microRNA mir-692 | 82 | 2371 | 347.5 | Reduced aligment | ECs |  |
| RF00876 | microRNA mir-684 | 89 | 2250 | 298.0 | Reduced aligment | ECs |  |
| RF00885 | microRNA MIR821 | 276 | 2232 | 158.7 | Reduced aligment | ECs |  |
| RF00906 | microRNA MIR1122 | 115 | 927 | 104.8 | Reduced aligment | ECs |  |
| RF00989 | microRNA mir-492 | 117 | 1793 | 99.5 | Reduced aligment | ECs |  |
| RF01016 | microRNA mir-584 | 121 | 408 | 146.3 | Reduced aligment | ECs |  |
| RF01045 | microRNA mir-544 | 106 | 1634 | 147.9 | Reduced aligment | ECs |  |
| RF01051 | Cyclic di-GMP-I riboswitch | 88 | 1990 | 659.7 | Reduced aligment | ECs |  |
| RF01055 | Moco (molybdenum cofactor) riboswitch | 138 | 1080 | 238.8 | Reduced aligment | ECs |  |
| RF01057 | S-adenosyl-L-homocysteine riboswitch | 78 | 419 | 125.5 | Reduced aligment | ECs |  |
| RF01059 | microRNA mir-598 | 97 | 4886 | 1276.7 | Reduced aligment | ECs |  |
| RF01063 | microRNA mir-324 | 67 | 2918 | 589.8 | Reduced aligment | ECs |  |
| RF01068 | mini-ykkC RNA motif | 48 | 932 | 91.1 | Reduced aligment | ECs |  |
| RF01159 | Small nucleolar RNA U18 | 72 | 171 | 48.7 | Reduced aligment | ECs |  |
| RF01185 | Small nucleolar RNA snR75 | 87 | 221 | 43.5 | Reduced aligment | ECs |  |
| RF01225 | Small nucleolar RNA ACA64 | 128 | 334 | 93.7 | Reduced aligment | ECs |  |
| RF01233 | Small nucleolar RNA U109 | 136 | 426 | 81.5 | Reduced aligment | ECs |  |
| RF01394 | isrK Hfq binding RNA | 78 | 1476 | 169.0 | Reduced aligment | ECs |  |
| RF01413 | microRNA miR-430 | 88 | 2185 | 77.5 | Reduced aligment | ECs |  |
| RF01417 | Retroviral 3'UTR stability element | 290 | 21039 | 1314.9 | Reduced aligment | ECs |  |
| RF01419 | Antisense RNA which regulates isiA expression | 188 | 2410 | 159.6 | Reduced aligment | ECs |  |
| RF01421 | small nucleolar RNA snoR114 | 88 | 206 | 79.5 | Reduced aligment | ECs |  |
| RF01492 | Listeria sRNA rli28 | 184 | 1124 | 120.0 | Reduced aligment | ECs |  |
| RF01497 | ALIL pseudoknot | 118 | 3080 | 141.3 | Reduced aligment | ECs |  |
| RF01656 | C. elegans sRNA ceN72-3_ceN74-2 | 70 | 181 | 50.8 | Reduced aligment | ECs |  |
| RF01688 | Actino-pnp RNA | 61 | 409 | 36.1 | Reduced aligment | ECs |  |
| RF01690 | Bacillaceae-1 RNA | 68 | 408 | 43.7 | Reduced aligment | ECs |  |
| RF01695 | C4 antisense RNA | 88 | 3693 | 462.2 | Reduced aligment | ECs |  |
| RF01699 | Clostridiales-1 RNA | 168 | 873 | 311.1 | Reduced aligment | ECs |  |
| RF01701 | Cyano-1 RNA | 70 | 437 | 81.4 | Reduced aligment | ECs |  |
| RF01704 | Downstream peptide RNA | 62 | 858 | 96.4 | Reduced aligment | ECs |  |
| RF01705 | Flavo-1 RNA | 77 | 959 | 397.0 | Reduced aligment | ECs |  |
| RF01717 | PhotoRC-II RNA | 95 | 568 | 99.5 | Reduced aligment | ECs |  |
| RF01725 | SAM-I/IV variant riboswitch | 99 | 734 | 201.5 | Reduced aligment | ECs |  |
| RF01726 | SAM-II long loop | 60 | 324 | 54.7 | Reduced aligment | ECs |  |
| RF01727 | SAM/SAH riboswitch | 50 | 88 | 25.8 | Reduced aligment | ECs |  |
| RF01731 | TwoAYGGAY RNA | 168 | 701 | 174.3 | Reduced aligment | ECs |  |
| RF01739 | glnA RNA | 54 | 1434 | 72.8 | Reduced aligment | ECs |  |
| RF01746 | mraW RNA | 108 | 342 | 80.6 | Reduced aligment | ECs |  |
| RF01747 | msiK RNA | 61 | 423 | 67.9 | Reduced aligment | ECs |  |
| RF01749 | pan motif | 92 | 400 | 98.6 | Reduced aligment | ECs |  |
| RF01750 | pfl RNA | 86 | 709 | 254.3 | Reduced aligment | ECs |  |
| RF01754 | radC RNA | 150 | 529 | 111.3 | Reduced aligment | ECs |  |
| RF01761 | wcaG RNA | 99 | 164 | 57.4 | Reduced aligment | ECs |  |
| RF01763 | ykkC-III RNA | 68 | 169 | 81.5 | Reduced aligment | ECs |  |
| RF01764 | yjdF RNA | 101 | 610 | 71.9 | Reduced aligment | ECs |  |
| RF01766 | cspA thermoregulator | 390 | 6179 | 1469.2 | Reduced aligment | ECs |  |
| RF01776 | RNA anti-toxin A | 90 | 642 | 59.7 | Reduced aligment | ECs |  |
| RF01787 | drz-agam-1 riboswitch | 117 | 5017 | 108.6 | Reduced aligment | ECs |  |
| RF01788 | drz-agam-2-2 riboswitch | 193 | 740 | 104.2 | Reduced aligment | ECs |  |
| RF01793 | ffh sRNA | 54 | 1092 | 38.8 | Reduced aligment | ECs |  |
| RF01794 | sok antitoxin | 157 | 3129 | 142.5 | Reduced aligment | ECs |  |
| RF01844 | SmY spliceosomal RNA | 81 | 172 | 69.4 | Reduced aligment | ECs |  |
| RF01847 | Plant small nucleolar RNA U3 | 222 | 572 | 152.5 | Reduced aligment | ECs |  |
| RF01849 | Alphaproteobacteria transfer-messenger RNA | 304 | 1712 | 224.9 | Reduced aligment | ECs |  |
| RF01850 | Betaproteobacteria transfer-messenger RNA | 201 | 615 | 137.7 | Reduced aligment | ECs |  |
| RF01853 | Mitochondrial DNA control region secondary structure A | 107 | 61513 | 198.2 | Reduced aligment | ECs |  |
| RF01854 | Bacterial large signal recognition particle RNA | 258 | 1662 | 368.7 | Reduced aligment | ECs |  |
| RF01855 | Plant signal recognition particle RNA | 306 | 687 | 164.4 | Reduced aligment | ECs |  |
| RF01867 | caulobacter sRNA CC2171 | 105 | 141 | 68.9 | Reduced aligment | ECs |  |
| RF01942 | microRNA mir-1937 | 120 | 9592 | 341.5 | Reduced aligment | ECs |  |
| RF01998 | Group II catalytic intron D1-D4-1 | 85 | 1726 | 320.4 | Reduced aligment | ECs |  |
| RF01999 | Group II catalytic intron D1-D4-2 | 119 | 594 | 136.3 | Reduced aligment | ECs |  |
| RF02003 | Group II catalytic intron D1-D4-4 | 140 | 360 | 93.7 | Reduced aligment | ECs |  |
| RF02004 | Group II catalytic intron D1-D4-5 | 199 | 555 | 136.0 | Reduced aligment | ECs |  |
| RF02005 | Group II catalytic intron D1-D4-6 | 234 | 666 | 289.0 | Reduced aligment | ECs |  |
| RF02033 | HNH endonuclease-associated RNA and ORF (HEARO) RNA | 128 | 458 | 178.0 | Reduced aligment | ECs |  |
| RF02076 | Gammaproteobacterial sRNA STnc100 | 207 | 1477 | 157.1 | Reduced aligment | ECs |  |
| RF02094 | mir-1803 microRNA precursor | 87 | 285 | 198.5 | Reduced aligment | ECs |  |
| RF02096 | mir-2973 microRNA precursor | 79 | 769 | 49.7 | Reduced aligment | ECs |  |
| RF02194 | Bacterial antisene RNA HPnc0260 | 155 | 792 | 259.4 | Reduced aligment | ECs |  |
| RF02221 | sRNA-Xcc1 | 88 | 774 | 113.5 | Reduced aligment | ECs |  |
| RF02223 | Proteobacterial sRNA sX4 | 148 | 210 | 111.4 | Reduced aligment | ECs |  |
| RF02228 | Xanthomonas sRNA sX9 | 77 | 1935 | 155.4 | Reduced aligment | ECs |  |
| RF02271 | TUC338 | 181 | 857 | 384.9 | Reduced aligment | ECs |  |
| RF02276 | Hammerhead ribozyme (type II) | 69 | 156 | 38.4 | Reduced aligment | ECs |  |